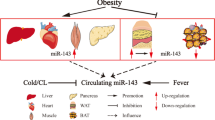
Key Points
-
MicroRNAs (miRNAs) are important for fat cell formation (adipogenesis) and for regulating the metabolic and endocrine functions of these cells
-
Obesity influences the expression of miRNAs in adipose tissue, but altered expression of only a few of these miRNAs has been experimentally verified in humans
-
Regional variations in expression of miRNAs in human adipose tissues have been demonstrated
-
miRNAs signal through complex networks involving transcription factors, which has been demonstrated in the context of regulation of inflammation in human adipose tissue
-
Extracellular miRNAs have specific expression profiles in obesity
Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression and, therefore, biological processes in different tissues. A major function of miRNAs in adipose tissue is to stimulate or inhibit the differentiation of adipocytes, and to regulate specific metabolic and endocrine functions. Numerous miRNAs are present in human adipose tissue; however, the expression of only a few is altered in individuals with obesity and type 2 diabetes mellitus or are differentially expressed in various adipose depots. In humans, obesity is associated with chronic low-grade inflammation that is regulated by signal transduction networks, in which miRNAs, either directly or indirectly (through regulatory elements such as transcription factors), influence the expression and secretion of inflammatory proteins. In addition to their diverse effects on signalling, miRNAs and transcription factors can interact to amplify the inflammatory effect. Although additional miRNA signal networks in human adipose tissue are not yet known, similar regulatory circuits have been described in brown adipose tissue in mice. miRNAs can also be secreted from fat cells into the circulation and serve as markers of disturbed adipose tissue function. Given their role in regulating transcriptional networks, miRNAs in adipose tissue might offer tangible targets for treating metabolic disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Haslam, D. W. & James, W. P. Obesity. Lancet 366, 1197–1209 (2005).
Chen, L., Magliano, D. J. & Zimmet, P. Z. The worldwide epidemiology of type 2 diabetes mellitus—present and future perspectives. Nat. Rev. Endocrinol. 8, 228–236 (2012).
Prentice, A. M. The emerging epidemic of obesity in developing countries. Int. J. Epidemiol. 35, 93–99 (2006).
Bray, G. A. & Tartaglia, L. A. Medicinal strategies in the treatment of obesity. Nature 404, 672–677 (2000).
Rosen, E. D. & MacDougald, O. A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885–896 (2006).
Arner, E. & Arner, P. Health and obesity: not just skin deep. Science 342, 558–559 (2013).
Arner, P. Not all fat is alike. Lancet 351, 1301–1302 (1998).
Arner, P. & Langin, D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol. Metab. 25, 255–262 (2014).
Johnson, A. M. & Olefsky, J. M. The origins and drivers of insulin resistance. Cell 152, 673–684 (2013).
Turer, A. T. & Scherer, P. E. Adiponectin: mechanistic insights and clinical implications. Diabetologia 55, 2319–2326 (2012).
Hotamisligil, G. S. Inflammation and metabolic disorders. Nature 444, 860–867 (2006).
Rosen, E. D. & Spiegelman, B. M. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16, 145–171 (2000).
Arner, P. et al. Dynamics of human adipose lipid turnover in health and metabolic disease. Nature 478, 110–113 (2011).
Ryden, M., Andersson, D. P., Bernard, S., Spalding, K. & Arner, P. Adipocyte triglyceride turnover and lipolysis in lean and overweight subjects. J. Lipid Res. 54, 2909–2913 (2013).
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Nuutila, P. Brown adipose tissue thermogenesis in humans. Diabetologia 56, 2110–2112 (2013).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Mukherji, S. et al. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 43, 854–859 (2011).
Lu, J. & Clark, A. G. Impact of microRNA regulation on variation in human gene expression. Genome Res. 22, 1243–1254 (2012).
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011).
Huang, J. C. et al. Using expression profiling data to identify human microRNA targets. Nat. Methods 4, 1045–1049 (2007).
Ulitsky, I., Laurent, L. C. & Shamir, R. Towards computational prediction of microRNA function and activity. Nucleic Acids Res. 38, e160 (2010).
Gennarino, V. A. et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 22, 1163–1172 (2012).
Huang, G. T., Athanassiou, C. & Benos, P. V. mirConnX: condition-specific mRNA–microRNA network integrator. Nucleic Acids Res. 39, W416–W423 (2011).
Jayaswal, V., Lutherborrow, M., Ma, D. D. & Yang, Y. H. Identification of microRNA–mRNA modules using microarray data. BMC Genomics 12, 138 (2011).
Le Bechec, A. et al. MIR@NT@N: a framework integrating transcription factors, microRNAs and their targets to identify sub-network motifs in a meta-regulation network model. BMC Bioinformatics 12, 67 (2011).
Xu, J. et al. MiRNA–miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res. 39, 825–836 (2011).
Kuhn, D. E. et al. Experimental validation of miRNA targets. Methods 44, 47–54 (2008).
Xie, H., Lim, B. & Lodish, H. F. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58, 1050–1057 (2009).
Arner, E. et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 61, 1986–1993 (2012).
Heneghan, H. M., Miller, N., McAnena, O. J., O'Brien, T. & Kerin, M. J. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J. Clin. Endocrinol. Metab. 96, E846–E850 (2011).
Keller, P. et al. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 11, 7 (2011).
Martinelli, R. et al. miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 18, 2170–2176 (2010).
Meerson, A. et al. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNFα. Diabetologia 56, 1971–1979 (2013).
Ortega, F. J. et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 5, e9022 (2010).
Capobianco, V. et al. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J. Proteome Res. 11, 3358–3369 (2012).
Chen, L. et al. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Mol. Cell. Endocrinol. 393, 65–74 (2014).
Chou, W. W. et al. Decreased microRNA-221 is associated with high levels of TNFα in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell Physiol. Biochem. 32, 127–137 (2013).
Diawara, M. R. et al. Adaptive expression of microRNA-125a in adipose tissue in response to obesity in mice and men. PLoS ONE 9, e91375 (2014).
Oger, F. et al. Cell-specific dysregulation of microRNA expression in obese white adipose tissue. J. Clin. Endocrinol. Metab. 99, 2821–2833 (2014).
Dahlman, I. et al. Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor α. Diabetes 55, 1792–1799 (2006).
Klimcakova, E. et al. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: decreased metabolism and increased immune response. J. Clin. Endocrinol. Metab. 96, E73–E82 (2011).
Hilton, C., Neville, M. J. & Karpe, F. MicroRNAs in adipose tissue: their role in adipogenesis and obesity. Int. J. Obesity 37, 325–332 (2013).
Neville, M. J., Collins, J. M., Gloyn, A. L., McCarthy, M. I. & Karpe, F. Comprehensive human adipose tissue mRNA and microRNA endogenous control selection for quantitative real-time-PCR normalization. Obesity (Silver Spring) 19, 888–892 (2011).
Civelek, M. et al. Genetic regulation of human adipose microRNA expression and its consequences for metabolic traits. Hum. Mol. Genet. 22, 3023–3037 (2013).
Vohl, M. C. et al. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes. Res. 12, 1217–1222 (2004).
Kloting, N. et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE 4, e4699 (2009).
Yu, J. et al. Expression profiling of PPARγ-regulated microRNAs in human subcutaneous and visceral adipogenesis in both genders. Endocrinology 155, 2155–2165 (2014).
Rantalainen, M. et al. MicroRNA expression in abdominal and gluteal adipose tissue is associated with mRNA expression levels and partly genetically driven. PLoS ONE 6, e27338 (2011).
Honardoost, M., Sarookhani, M. R., Arefian, E. & Soleimani, M. Insulin resistance associated genes and miRNAs. Appl. Biochem. Biotechnol. 174, 63–80 (2014).
Rottiers, V. & Naar, A. M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012).
Wu, H. L. et al. The expression of the miR-25/93/106b family of micro-RNAs in the adipose tissue of women with polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 99, E2754–E2761 (2014).
Chen, Y. H. et al. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes 62, 2278–2286 (2013).
Lorente-Cebrian, S. et al. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNFα. PLoS ONE 9, e86800 (2014).
Lin, Y. Y. et al. KSRP and microRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol. Cell. Biol. 34, 2339–2349 (2014).
Kang, M. et al. Role of microRNA-21 in regulating 3T3-L1 adipocyte differentiation and adiponectin expression. Mol. Biol. Rep. 40, 5027–5034 (2013).
Shi, Z. et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERα expression in estrogen-induced insulin resistance. Endocrinology 155, 1982–1990 (2014).
Ferland-McCollough, D., Ozanne, S. E., Siddle, K., Willis, A. E. & Bushell, M. The involvement of microRNAs in type 2 diabetes. Biochem. Soc. Trans. 38, 1565–1570 (2010).
Maury, E. & Brichard, S. M. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol. Cell. Endocrinol. 314, 1–16 (2010).
Sonkoly, E. & Pivarcsi, A. microRNAs in inflammation. Int. Rev. Immunol. 28, 535–561 (2009).
Ge, Q., Brichard, S., Yi, X. & Li, Q. microRNAs as a new mechanism regulating adipose tissue inflammation in obesity and as a novel therapeutic strategy in the metabolic syndrome. J. Immunol. Res. 2014, 987285 (2014).
Hulsmans, M., De Keyzer, D. & Holvoet, P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB J. 25, 2515–2527 (2011).
Strum, J. C. et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 23, 1876–1884 (2009).
Zhuang, G. et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 125, 2892–2903 (2012).
Shi, C. et al. IL-6 and TNFα induced obesity-related inflammatory response through transcriptional regulation of miR-146b. J. Interferon Cytokine Res. 34, 342–348 (2014).
Zhu, L. et al. MiR-335, an adipogenesis-related microRNA, is involved in adipose tissue inflammation. Cell Biochem. Biophys. 68, 283–290 (2014).
Kim, C. et al. TNFα -induced miR-130 resulted in adipocyte dysfunction during obesity-related inflammation. FEBS Lett. 587, 3853–3858 (2013).
Ge, Q., Gerard, J., Noel, L., Scroyen, I. & Brichard, S. M. MicroRNAs regulated by adiponectin as novel targets for controlling adipose tissue inflammation. Endocrinology 153, 5285–5296 (2012).
Subedi, A. & Park, P. H. Autocrine and paracrine modulation of microRNA-155 expression by globular adiponectin in RAW 264.7 macrophages: involvement of MAPK/NF-κB pathway. Cytokine 64, 638–641 (2013).
Parra, P., Serra, F. & Palou, A. Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS ONE 5, e13005 (2010).
Lefterova, M. I. & Lazar, M. A. New developments in adipogenesis. Trends Endocrinol. Metab. 20, 107–114 (2009).
Oskowitz, A. Z. et al. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc. Natl Acad. Sci. USA 105, 18372–18377 (2008).
Mudhasani, R., Imbalzano, A. N. & Jones, S. N. An essential role for Dicer in adipocyte differentiation. J. Cell. Biochem. 110, 812–816 (2010).
McGregor, R. A. & Choi, M. S. microRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 11, 304–316 (2011).
Ling, H. et al. The physiological and pathophysiological roles of adipocyte miRNAs. Biochem. Cell Biol. 91, 195–202 (2013).
Peng, Y. et al. MicroRNAs: Emerging roles in adipogenesis and obesity. Cell. Signal. 26, 1888–1896 (2014).
Alexander, R., Lodish, H. & Sun, L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin. Ther. Targets 15, 623–636 (2011).
Son, Y. H., Ka, S., Kim, A. Y. & Kim, J. B. Regulation of adipocyte differentiation via microRNAs. Endocrinol. Metab. (Seoul) 29, 122–135 (2014).
Lee, Y. S. & Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21, 1025–1030 (2007).
Sun, T., Fu, M., Bookout, A. L., Kliewer, S. A. & Mangelsdorf, D. J. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol. 23, 925–931 (2009).
Kim, Y. J., Hwang, S. J., Bae, Y. C. & Jung, J. S. MiR-21 regulates adipogenic differentiation through the modulation of TGFβ signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 27, 3093–3102 (2009).
Huang, S. et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 21, 2531–2540 (2012).
Karbiener, M. et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARγ. Biochem. Biophys. Res. Commun. 390, 247–251 (2009).
Lin, Q., Gao, Z., Alarcon, R. M., Ye, J. & Yun, Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 276, 2348–2358 (2009).
Sun, F. et al. Characterization of function and regulation of miR-24-1 and miR-31. Biochem. Biophys. Res. Commun. 380, 660–665 (2009).
Tang, Y. F. et al. Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS 13, 331–336 (2009).
Lee, E. K. et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor γ expression. Mol. Cell Biol. 31, 626–638 (2011).
Yang, Z. et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 20, 259–267 (2011).
Guo, Y., Chen, Y., Zhang, Y., Chen, L. & Mo, D. Up-regulated miR-145 expression inhibits porcine preadipocytes differentiation by targeting IRS1. Int. J. Biol. Sci. 8, 1408–1417 (2012).
Liu, S., Yang, Y. & Wu, J. TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochem. Biophys. Res. Commun. 414, 618–624 (2011).
Peng, Y. et al. MiR-224 impairs adipocyte early differentiation and regulates fatty acid metabolism. Int. J. Biochem. Cell Biol. 45, 1585–1593 (2013).
Bork, S. et al. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J. Cell Physiol. 226, 2226–2234 (2011).
Kinoshita, M. et al. Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol. Endocrinol. 24, 1978–1987 (2010).
Wang, Q. et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl Acad. Sci. USA 105, 2889–2894 (2008).
Esau, C. et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279, 52361–52365 (2004).
Chen, L. et al. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5–ERK5 signaling. Sci. Rep. 4, 3819 (2014).
Karbiener, M. et al. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biol. 8, 850–860 (2011).
Zaragosi, L. E. et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 12, R64 (2011).
Kim, S. Y. et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARγ expression. Biochem. Biophys. Res. Commun. 392, 323–328 (2010).
Zhang, J. F. et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol. Biol. Cell 22, 3955–3961 (2011).
Skarn, M. et al. Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev. 21, 873–883 (2012).
Peirce, V., Carobbio, S. & Vidal-Puig, A. The different shades of fat. Nature 510, 76–83 (2014).
Beranger, G. E. et al. In vitro brown and “brite”/“beige” adipogenesis: human cellular models and molecular aspects. Biochim. Biophys. Acta 1831, 905–914 (2013).
Trajkovski, M. & Lodish, H. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol. Metab. 24, 442–450 (2013).
Karbiener, M. et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 32, 1578–1590 (2014).
Mori, M., Nakagami, H., Rodriguez-Araujo, G., Nimura, K. & Kaneda, Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 10, e1001314 (2012).
Chen, Y. et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 4, 1769 (2013).
Liu, W. et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 9, e1003626 (2013).
Martinez, N. J. & Walhout, A. J. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays 31, 435–445 (2009).
Schadt, E. E. Molecular networks as sensors and drivers of common human diseases. Nature 461, 218–223 (2009).
Sieberts, S. K. & Schadt, E. E. Moving toward a system genetics view of disease. Mamm. Genome 18, 389–401 (2007).
Sato, F., Tsuchiya, S., Meltzer, S. J. & Shimizu, K. MicroRNAs and epigenetics. FEBS J. 278, 1598–1609 (2011).
Inui, M., Martello, G. & Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 11, 252–263 (2010).
Hagen, J. W. & Lai, E. C. microRNA control of cell–cell signaling during development and disease. Cell Cycle 7, 2327–2332 (2008).
Ichimura, A., Ruike, Y., Terasawa, K. & Tsujimoto, G. miRNAs and regulation of cell signaling. FEBS J. 278, 1610–1618 (2011).
Herranz, H. & Cohen, S. M. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24, 1339–1344 (2010).
Gomes, C. P. et al. A review of computational tools in microRNA discovery. Front. Genet. 4, 81 (2013).
Meyer, S. U. et al. Posttranscriptional regulatory networks: from expression profiling to integrative analysis of mRNA and microRNA data. Methods Mol. Biol. 1160, 165–188 (2014).
Tsang, J. S., Ebert, M. S. & van Oudenaarden, A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell 38, 140–153 (2010).
Tsang, J., Zhu, J. & van Oudenaarden, A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26, 753–767 (2007).
Chavali, S. et al. MicroRNAs act complementarily to regulate disease-related mRNA modules in human diseases. RNA 19, 1552–1562 (2013).
Kulyté, A. et al. Additive effects of miRNAs and transcription factors on CCL2 production in human white adipose tissue. Diabetes 63, 1248–1258 (2014).
Kim, V. N., Han, J. & Siomi, M. C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 (2009).
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108, 5003–5008 (2011).
Turchinovich, A., Weiz, L., Langheinz, A. & Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011).
Mathivanan, S., Ji, H. & Simpson, R. J. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 (2010).
Li, L. et al. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS ONE 7, e46957 (2012).
Huang, X. et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14, 319 (2013).
Williams, Z. et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl Acad. Sci. USA 110, 4255–4260 (2013).
Blondal, T. et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6 (2013).
Hu, Z. et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28, 1721–1726 (2010).
Ogawa, R. et al. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem. Biophys. Res. Commun. 398, 723–729 (2010).
Muller, G., Schneider, M., Biemer-Daub, G. & Wied, S. Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell Signal 23, 1207–1223 (2011).
Wang, Y. C. et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 56, 2275–2285 (2013).
Deng, Z. B. et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 58, 2498–2505 (2009).
Koeck, E. S. et al. Adipocyte exosomes induce transforming growth factor β pathway dysregulation in hepatocytes: a novel paradigm for obesity-related liver disease. J. Surg. Res. 192, 268–275 (2014).
Guay, C. & Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 9, 513–521 (2013).
Rome, S. Are extracellular microRNAs involved in type 2 diabetes and related pathologies? Clin. Biochem. 46, 937–945 (2013).
Karolina, D. S. et al. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 97, E2271–E2276 (2012).
Ortega, F. J. et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 37, 1375–1383 (2014).
Zampetaki, A. et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 107, 810–817 (2010).
Ortega, F. J. et al. Targeting the circulating microRNA signature of obesity. Clin. Chem. 59, 781–792 (2013).
Wang, Y. T., Tsai, P. C., Liao, Y. C., Hsu, C. Y. & Juo, S. H. Circulating microRNAs have a sex-specific association with metabolic syndrome. J. Biomed. Sci. 20, 72 (2013).
Prats-Puig, A. et al. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 98, E1655–E1660 (2013).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 438, 685–689 (2005).
Esau, C. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 (2006).
Elmen, J. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008).
Wahid, F., Shehzad, A., Khan, T. & Kim, Y. Y. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta 1803, 1231–1243 (2010).
US National Institutes of Health. ClinicalTrials.gov [online], (2014).
van Rooij, E., Purcell, A. L. & Levin, A. A. Developing microRNA therapeutics. Circ. Res. 110, 496–507 (2012).
Cao, Y. Adipose tissue angiogenesis as a therapeutic target for obesity and metabolic diseases. Nat. Rev. Drug Discov. 9, 107–115 (2010).
Czech, M. P., Aouadi, M. & Tesz, G. J. RNAi-based therapeutic strategies for metabolic disease. Nat. Rev. Endocrinol. 7, 473–484 (2011).
Li, Z. & Rana, T. M. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 13, 622–638 (2014).
Xu, G. et al. Modulation of hsa-miR-26b levels following adipokine stimulation. Mol. Biol. Rep. 40, 3577–3582 (2013).
Chen, T. et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc. Res. 83, 131–139 (2009).
Estep, M. et al. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 32, 487–497 (2010).
Zhu, L. et al. FFAs and adipokine-mediated regulation of hsa-miR-143 expression in human adipocytes. Mol. Biol. Rep. 40, 5669–5675 (2013).
Hulsmans, M., Van Dooren, E., Mathieu, C. & Holvoet, P. Decrease of miR-146b-5p in monocytes during obesity is associated with loss of the anti-inflammatory but not insulin signaling action of adiponectin. PLoS ONE 7, e32794 (2012).
Huang, R. S., Hu, G. Q., Lin, B., Lin, Z. Y. & Sun, C. C. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J. Investig. Med. 58, 961–967 (2010).
Urbich, C., Kuehbacher, A. & Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 79, 581–588 (2008).
Pritchard, C. C., Cheng, H. H. & Tewari, M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 13, 358–369 (2012).
Acknowledgements
P.A. has received funding from the Swedish Research Council, the Swedish Diabetes Foundation and the Diabetes Program at Karolinska Institutet. A.K. has received funding from the Åke Wiberg Foundation and Tore Nilsson Foundation.
Author information
Authors and Affiliations
Contributions
P.A. and A.K. contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
P.A. has received funding from the Novo Nordisk Foundation, which is a non-profit organization fully independent of the pharmaceutical company Novo Nordisk Ltd. A.K. declares no competing interests.
Rights and permissions
About this article
Cite this article
Arner, P., Kulyté, A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 11, 276–288 (2015). https://doi.org/10.1038/nrendo.2015.25
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrendo.2015.25
This article is cited by
-
Adipo-oncology: adipocyte-derived factors govern engraftment, survival, and progression of metastatic cancers
Cell Communication and Signaling (2024)
-
Lean adipose tissue macrophage derived exosome confers immunoregulation to improve wound healing in diabetes
Journal of Nanobiotechnology (2023)
-
Identification of novel circulating miRNAs biomarkers for healthy obese and lean children
BMC Endocrine Disorders (2023)
-
Significant role of some miRNAs as biomarkers for the degree of obesity
Journal of Genetic Engineering and Biotechnology (2023)
-
MicroRNAs: a crossroad that connects obesity to immunity and aging
Immunity & Ageing (2022)